Case Study Tutorials¶
Here you can find a simple example on how to run a clustering job using BitQT as well as how to visualize the clusters using a VMD plugin.
Clustering a MD¶
Note
We included an example folder where you can find the topology and trajectory files we have used in this section.
As we already mentioned, the only required argument for BitQT is the trajectory file. We will use the binary dcd file aligned_original_tau_6K.dcd. As dcd format does not contain any topological information, it is necessarty to pass the -top argument with an appropiate topology file. In this case, we will be using the PDB formatted file aligned_tau.pdb.
Then you can run
$ bitqt -top examples/aligned_tau.pdb -traj examples/aligned_original_tau_6K.dcd -sel all -cutoff 4 -odir 6K_4
After succesful termination, BitQT will produce some output files to the specified folder 6K_4:
A cluster_statistics.txt file containing clusterID, cluster_size, and its percentage from the total frames analyzed
A frames_statistics.txt file containing every frameID and its clusterID.
A file.log to visualize all the clusters via VMD plugin as discussed in the next section.
Visualizing Clusters in VMD¶
BitQT produces a file.log that contains cluster frames in the NMRcluster format. This can be visualized in VMD using the clustering plugin (see Installation).
Figure 1 shows the main window of the plugin and the steps you should follow:
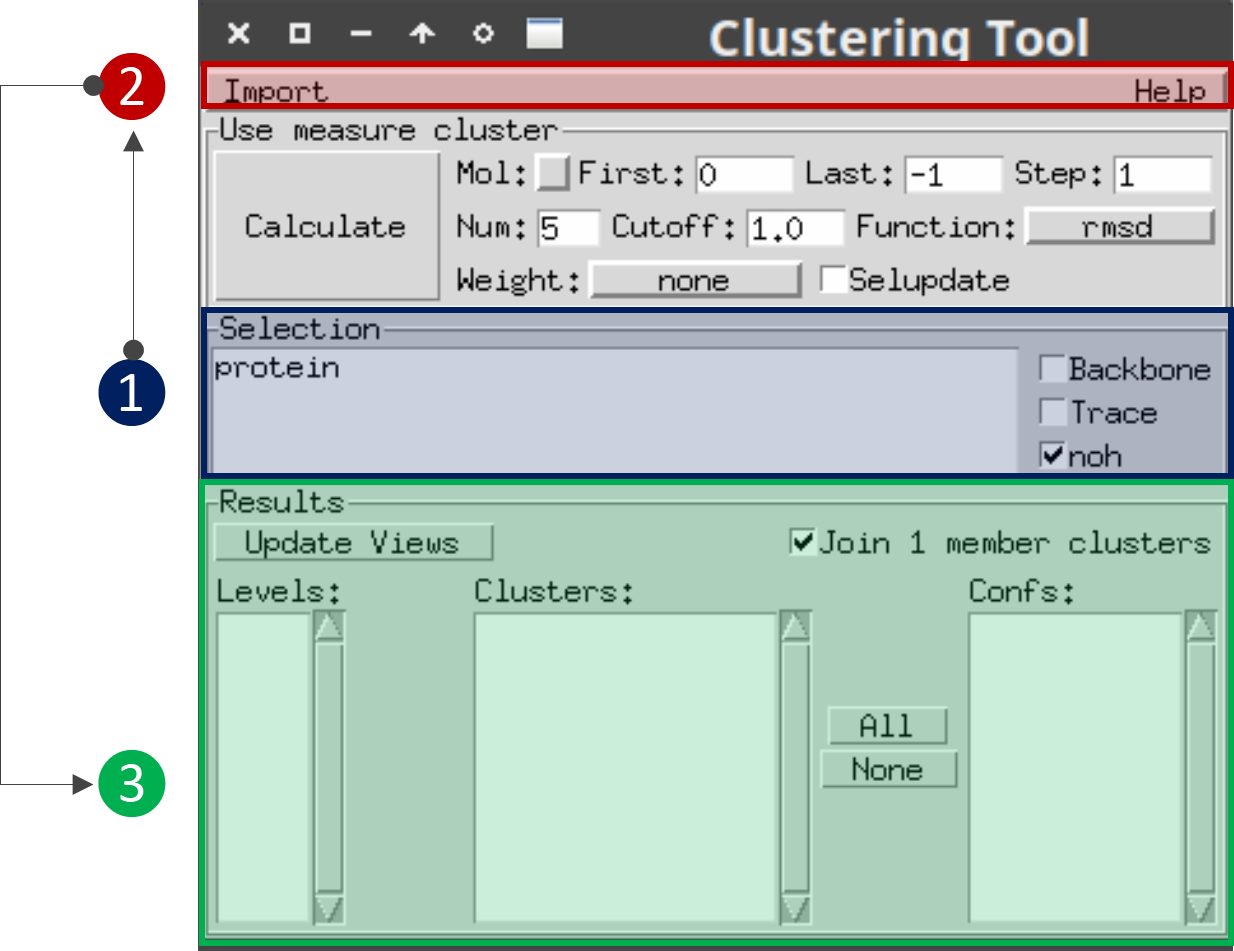
Figure 1: Main window of the VMD clustering plugin¶
Selection section: Here you can define the selection of atoms that you would like to visualize.
Import section: After loading in VMD the topology and trajectory files that you used to run BitQT, go to the Import button of the plugin, select NMRcluster option and navigate to the file.log produced by BitQT.
Results section: Here you can select which clusters to visualize. Note that through the standard VMD commands, you can change the representations and customize the visualization as you want.
Do not change any of the parameters from the Use measure cluster section. As it indicates, these are for triggering the internal measure cluster command of VMD that does not implement QT.
Figure 2 shows a loaded example. Note that only the backbone of clusters 1 and 4 have been selected.
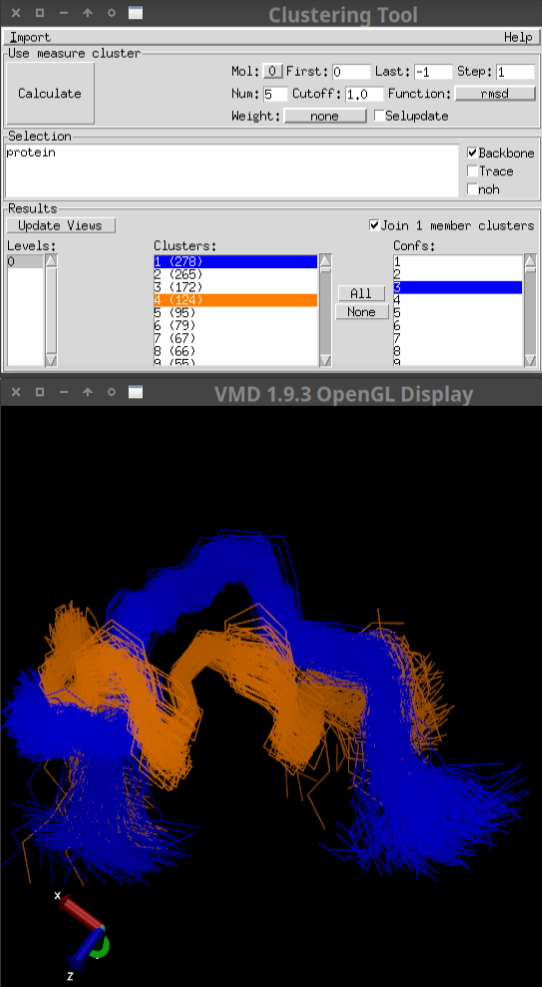
Figure 2: Visualization example¶